
Ralf Jauch Group Leader Dr. Ralf Jauch studied Biology, Psychology and Archeology in Jena (Germany) and Manchester (UK) before doing his PhD in a molecular biology program from 2001-2005 at the Max-Planck-Institut für biophyskalische Chemie in Göttingen, Germany. In 2006 he got attracted to Singapore to join the recently established Genome Institute of Singapore (GIS, part of the A*STAR family) for postdoctoral training. From 2008-2013 he continued at the GIS as a research scientist and drove an interdisciplinary program at the interface of structural biochemistry, genomics and stem cell biology. In 2013 he joined the GIBH as principle investigator and carries out translation research within the DDP as well as academic projects within the stem cell institute. Research: 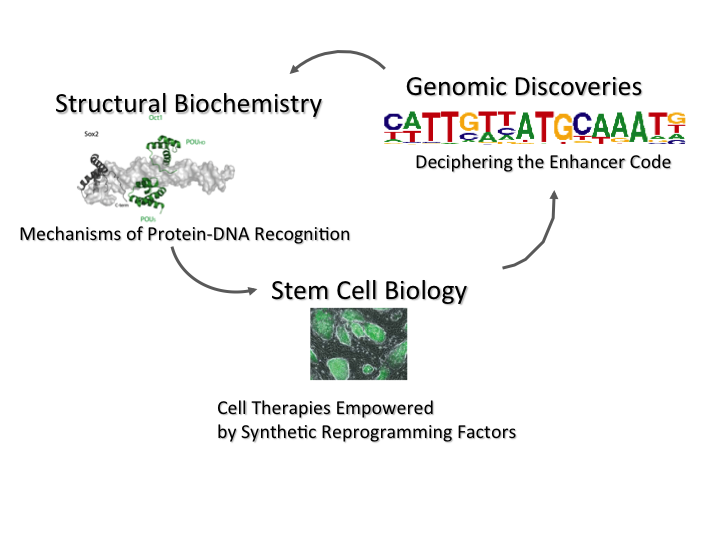
Our interest is to decipher the ‘enhancer code’, that is we wish to understand how regulatory information is genetically hard-wired in the genome. The ‘enhancer code’ determines when and where genes are expressed in our bodies and hence underlies the development of individuals, of species during evolution as well as of tumors when cancers arise. This code is ‘read’ by transcription factor proteins (TFs) that collaborate to trigger cell fate decisions. We use a combination of genomics (bioinformatics and next generation sequencing), structural biochemistry and stem stem cell biology. Goal of our work is to gain fundamental insights into genome regulation and protein-DNA recognition. We hope to translate these insights to generate synthetic factors to optimize cellular reprogramming to obtain cells suitable for replacement therapies. We also attempt to develop small-molecule as well as nucleic acid based drugs that target TFs and other genome regulators. Interested individuals are invited to submit their resumes to join our team for studentships, research associate and postdoctoral positions.
Publications (pubmed link): - Aksoy I. *, Jauch R. *(* shared first author), Eras V., Bin AC., Chen J., Divakar U., Ng CK., Kolatkar PR., Stanton LW. (2013) Sox transcription factors require selective interactions with Oct4 and specific transactivation functions to mediate reprogramming. Stem Cells 20 Aug [Epub]
- Jauch, R. and Kolatkar, P.R. (2013). What makes a pluripotency reprogramming factor? Curr Mol Med May 2
- Aksoy, I.*, Jauch, R. *(* shared first author), Jiaxuan, C., Dyla, M., Bogu, G.K., Divakar, U., Ng, C.K., Yi, R.T., Hutchins, A.P., Kolatkar, P.R. et al. (2013) Oct4 switches partnering from Sox2 to Sox17 and reinterprets the enhancer code to specify primitive endoderm. EMBO J, Apr 3;32(7):938-53. – highlighted in “Have you seen section”
- Esch, D., Vahokoski J., Goves, M.R., Pogenberg, V., Cojocaru V., Bruch, H., Han, D., Drexler, H.C.A., Arauzo-Bravo, M.J., Ng, C.K.L., Jauch, R., Wilmanns, M., Schöler,H.R. (2013) A unique Oct4 interface is crucial for reprogramming to pluripotency. Nature Cell Biol, 2013 Feb 3;15(3):295-301.
- Sun, W., Hu, X., Lim, M.K., Ng, C.K., Choo, S.H., Kolatkar, P.R., Jauch, R. (corresponding author) and Prabhakar, S. (2013) Estimating protein-DNA binding energies from in vivo binding profiles. Nucl Acids Res, 2013 Apr 16
- Jankowski, A., Szczurek, E., Jauch R., Tiuryn, J., Prabhakar, S.(2013) Comprehensive Prediction in 78 human cell lines reveals rigidity and compactness of transcription factor dimmers. Genome Research, 2013 Apr 3 Epub
- Hutchins, A.P., Choo, S.H., Misri, T., Woon, C., Jauch, R. and Robson, P. (2012) Co-motif discovery identifies an Esrrb-Sox2-DNA ternary complex as a mediator of transcriptional differences between mouse embryonic and epiblast stem cells. Stem Cells, Feb;31(2):269-81.
- Hutchins, A.P., Diez, Diego; Takahashi, Y; Ahmad, S,; Jauch, R.; Tremblay, M.; Miranda-Saavedra, D. Differential co-factor recruitment determines STAT3's cell type-independent and cell type-specific functions. Nucleic Acids Res, 2013 Feb 1;41(4):2155-70
- Yu, L., Pugalenthi, G., Hong, F., Sizun, J., Jauch, R., Stanton, L.W. and Kolatkar, P.R. (2012) Structural Analysis and dimerization profile of the SCAN domain of the pluripotency factor Zfp206 Nucleic Acids Res, 2012 Sep 1;40(17):8721-32
- Johnson, R., Richter, N., Bogu, G.K., Choo, S.H., Jauch, R. and Stanton, L.W. (2012) A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target Genes. PLoS Genet 2012 April; 8(4)
- Ng, C.K.L., Prabhakar, S Kolatkar, P.R. Jauch, R. (2012). Deciphering the Sox-Partner code by quantitative cooperativity measurements. Nucleic Acids Res 2012 Jun 1;40(11):4933-41
- Jauch, R. (corresponding author) Ng, C.K.L., Narasimhan, K, Kolatkar, P.R.. (2011). Crystal structure of the Sox4 HMG/DNA complex suggests a mechanism for the positional interdependence in DNA recognition. Biochem J 443(1):39-47
- Jauch, R. Hutchins, A., Aksoy, I., Ng, C.K.L., Xian, X.F., Palasingam, P., Robson, P., Stanton, W., and Kolatkar, P.R. (2011). Conversion of Sox17 into a Pluripotency Reprogramming Factor by Re-engineering its Association with Oct4 on DNA. Stem Cells Jun;29(6):940-51 Highlighted by Faculty1000
- Baburajendran, N.*, Jauch, R*, (*equal contribution, corresponding author)., Tan, C and Kolatkar, P.R. (2011). Structural basis for DNA recognition by constitutive Smad4 MH1 dimers. Nucleic Acids Res. Oct;39(18):8213-22
- Narasimhan, K., Pillay, S., Bin Ahmad, N.R., Bikadi, Z., Hazai, E., Yan, L., Kolatkar, P.R., Pervushin, K.and Jauch, R. (2011) Identification of a Polyoxometalate Inhibitor of the DNA Binding Activity of Sox2. ACS Chem Biol. Jun 17;6(6):573-81
- Jauch R (corresponding author), Choo SH, Ng CK, et al. Crystal structure of the dimeric Oct6 (POU3f1) POU domain bound to palindromic MORE DNA. Proteins. 2011. 79(2):674-7
- Baburajendran, N., Palasingam, P., Narasimhan, K., Sun, W., Prabhakar, S., Jauch, R (corresponding author)., and Kolatkar, P.R. (2010). Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-{beta} effectors. Nucleic Acids Res. Jun;38(10):3477-88
- Johnson, R., Richter, N., Jauch, R., Gaughwin, P.M., Zuccato, C., Cattaneo, E., and Stanton, L.W. (2010). The Human Accelerated Region 1 noncoding RNA is repressed by REST in Huntington's disease. Physiol Genomics.
- Palasingam, P*., Jauch, R.*,(*equal contribution, corresponding author), Ng, C.K. & Kolatkar, P.R. The structure of Sox17 bound to DNA reveals a conserved bending topology but selective protein interaction platforms. J Mol Biol. 2009 388, 619-30
- Johnson, R., Samuel, J., Ng, C.K., Jauch, R., Stanton, L.W., and Wood, I.C. (2009). Evolution of the vertebrate gene regulatory network controlled by the transcriptional repressor REST. Mol Biol Evol 26, 1491-1507.
- Baburajendran, N., Palasingam, P., Ng, C.K., Jauch, R. and Kolatkar, P.R. (2009) Crystal optimization and preliminary diffraction data analysis of the Smad1 MH1 domain bound to a palindromic SBE DNA element. Acta Crystallogr Sect F Struct Biol Cryst Commun, 65, 1105-1109.
- Ng, C.K., Palasingam, P., Venkatachalam, R., Baburajendran, N., Cheng, J., Jauch, R. and Kolatkar, P.R. (2008) Purification, crystallization and preliminary X-ray diffraction analysis of the HMG domain of Sox17 in complex with DNA. Acta Crystallogr Sect F Struct Biol Cryst Commun, 64, 1184-1187.
- Jauch R, Ng CKL, Saikatendu KS, Stevens RC, Kolatkar PR. Crystal Structure and DNA Binding of the Homeodomain of the Stem Cell Transcription Factor Nanog. J Mol Biol. 2008; 376(3): 758-70.
- Jauch R, Yeo HC, Kolatkar PR, and Clarke ND. Assessment of CASP7 structure predictions for template free targets. Proteins. 2007; 69 Suppl 8:57-67
- Jauch, R., Cho, MK, Jakel, S, Netter, C, Schreiter, K, Aicher, B, Zweckstetter, M, Jackle, H and Wahl, MC. Mitogen-activated Protein Kinases Interacting Kinases are Autoinhibited by a Reprogrammed Activation Segment. Embo J. 2006; 25, 4020-4032.
-
-
- Jauch R, Jäkel S, Netter C, Schreiter K, Aicher B, Jäckle H and Wahl MC. Structures of Mnk-2 Reveal an Inhibitory Conformation and a Zinc-Binding Site. Structure. 2005;(13): 1559-1568.
-
-
- Jauch R, Humm A, Huber R, Wahl MC. Structures of E. coli NAD Synthetase with Substrates and Product Reveal Mechanistic Rearrangements. J Biol Chem. 2005; 280: 15131 – 15140
- Jauch R, Bourenkov GP, Chung HR, Urlaub H, Reidt U, Jackle H, Wahl MC. The Zinc Finger-Associated Domain of the Drosophila Transcription Factor grauzone is a Novel Zinc-Coordinating Protein-Protein Interaction Module. Structure. 2003;(11):1393-402.
Granted Patents: Jauchr R, Kolatkar PR, Aksoy I, Stanton LW. Mutant Sox Proteins and methods of inducing pluripotency. No. 61/272,793, filed on November 4, 2009 Jauch R, Wahl MC et al. Crystallographic Structure of Mnk2 and Mnk1.(EP 050198899.3). assignee: DeveloGen AG and Max-Planck-Gesellschaft. filing date 13.9.2005
|