GIBH Develope a New Tool CACIMAR for Analyzing Cross-Species Single-Cell Sequencing Data
Recently, WANG Jie's research group from the Guangzhou Institutes of Biomedicine and Health (GIBH), the Chinese Academy of Sciences, developed a new computational tool named CACIMAR (Cross-species Analysis of Cell Identities, Markers, Regulations) for analyzing cross-species single-cell RNA sequencing (scRNA-seq) data. This tool can reveal the evolutionary conservation of cell types, markers, intracellular regulations, and intercellular interactions across species. The research has been published in Briefings in Bioinformatics under the title "CACIMAR: cross-species analysis of cell identities, markers, regulations, and interactions using single-cell RNA sequencing data".
Species show evolutionary conservation not only at the gene level, but also at the aspects of cell type, gene expression, gene regulation, and intercellular interaction. Single-cell RNA sequencing technology has been widely used to study single-cell transcriptomes across different species, providing new opportunities for studying evolutionary conservation at the cellular level. Existing algorithms can identify evolutionarily conserved cell types by analyzing cross-species scRNA-seq data, such as Seurat and SAMap. However, these methods determine conserved cell types through cross-species single-cell clustering, and their accuracy depends largely on the ability to correct cross-species batch effects. In performing clustering analysis on evolutionarily distant species, these methods encounter an obvious limitation. Moreover, no method is available to identify the evolutionary conservation of intercellular interactions.
To address the above issues, the research group developed a new R software package called CACIMAR to analyze cross-species scRNA-seq data. Conservation analysis in CACIMAR primarily consists of three steps. Firstly, CACIMAR identified cell types, markers, intracellular regulations, and intercellular interactions based on single-cell clustering analysis within each species. This strategy does not require cross-species single-cell clustering, effectively avoiding cross-species batch effects. Secondly, CACIMAR calculated new conservation scores based on statistical powers and cross-species homology of markers to identify conserved cell types. In addition, weighted sum models of the features were also built to measure the conservation of intracellular regulations and intercellular interactions, respectively. In the last step, conserved or species-specific cell types, intracellular regulations, and intercellular interactions across species were determined based on the calculated conservation scores. Using publicly available scRNA-seq data from mice, zebrafish, and chick during retinal regeneration, CACIMAR effectively identified conserved markers, cell types, intracellular regulations, and intercellular interactions in the retina among mice, zebrafish, and chick.
In summary, CACIMAR provided a new algorithm to overcome the challenges of existing methods for cross-species scRNA-seq data analysis, especially in identifying cross-species conserved and specific cell types, regulations, and interactions. Utilizing this algorithm, researchers can analyze the evolutionary conservation across various species at both cellular and molecular levels, providing a new perspective for understanding the mechanism of species evolution.
The research was funded by the National Natural Science Foundation of China, National Key Research and Development Project of China, and Natural Science Foundation of Guangdong Province.
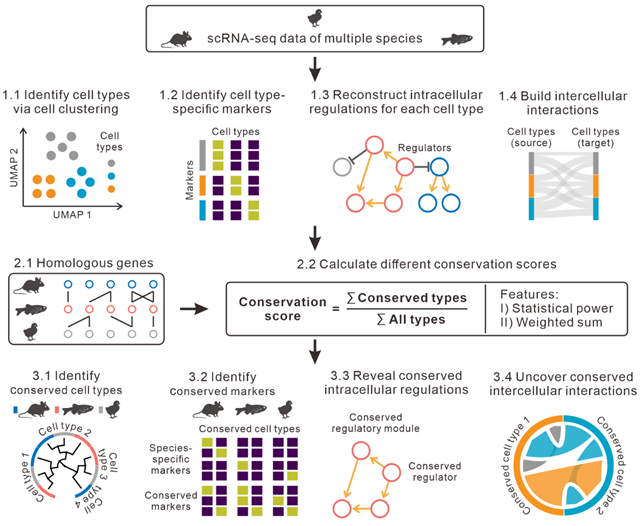
Schematic diagram of CACIMAR for cross-species scRNA-seq data analysis (Image by GIBH)
Contacts:
Jie Wang, Ph.D., Principle Investigator;
Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, China, 510530.
Email: wang_jie@gibh.ac.cn