Research Progress
Scientists reveal function of RNA m6A readers during somatic reprogramming
Date:Sep 09, 2020
On September 8, Cell Reports journal published a latest research titled “YTHDF2/3 are required for somatic reprogramming through different RNA deadenylation pathways” from Jiekai Chen’s group in Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences. This study revealed that the binding proteins YTHDF2 and YTHDF3 of m6A methylation on RNAs regulated the degradation of somatic genes synergistically through different RNA degradation pathways, and promoted the MET (mesenchymal-to-epithelial transition) process during somatic cell reprogramming.
m6A (N6-methyladenosine), one of the most common and abundant chemical modifications in eukaryotic mRNA, is regulated by methyltransferase complex (METTL3, METTL14, WTAP, etc.), demethylase (FTO and ALKBH5) and binding proteins (YTHDF1/2/3, YTHDC1/2). It is involved in the important physiological processes of stem cell fate determination and embryonic development1,2. Previous studies have shown that m6A plays an important role in stem cell self-renewal and differentiation and somatic cell reprogramming. However, studies under different conditions have different results3-6. These contradictory results may be due to the fact that m6A has a lot of complex functions, and these studies are focused on the m6A methylase METTL3, which will affect the level of m6A on the whole transcriptome, resulting in many disorders of m6A mediated functions.
m6A (N6-methyladenosine), one of the most common and abundant chemical modifications in eukaryotic mRNA, is regulated by methyltransferase complex (METTL3, METTL14, WTAP, etc.), demethylase (FTO and ALKBH5) and binding proteins (YTHDF1/2/3, YTHDC1/2). It is involved in the important physiological processes of stem cell fate determination and embryonic development1,2. Previous studies have shown that m6A plays an important role in stem cell self-renewal and differentiation and somatic cell reprogramming. However, studies under different conditions have different results3-6. These contradictory results may be due to the fact that m6A has a lot of complex functions, and these studies are focused on the m6A methylase METTL3, which will affect the level of m6A on the whole transcriptome, resulting in many disorders of m6A mediated functions.
In order to study the role of m6A in somatic reprogramming, this study focused on the relatively single function of the m6A binding protein YTHDF protein family. The results showed that knockdown of Ythdf2 or Ythdf3, but not Ythdf1, could inhibit the efficiency of somatic cell reprogramming, and the inhibition was m6A dependent. Further studies showed that YTHDF2 regulated somatic cell reprogramming by recruiting CCR4-NOT deadenylation complex, while YTHDF3 interacted with PAN3, recruiting PAN2-PAN3 complex to regulate mRNA degradation. During reprogramming, two different mRNA degradation pathways, YTHDF2-CCR4-NOT and YTHDF3-PAN2-PAN3, promoted the rapid clearance of somatic gene mRNA, which is conducive to the transformation of gene expression network from somatic cells to pluripotent stem cells. When one of these pathways is damaged, it will lead to prolong the lifetime of somatic gene mRNA, which will damage reprogramming.
In addition, we screened the genes with prolonged lifetime of mRNA after Ythdf2/3 knockdown, and found that TEAD2, the effector of Hippo signaling pathway, is an important target gene of YTHDF2/3. TEAD2 bound to EMT (epithelial-to-mesenchymal transition) related genes in the early stage of reprogramming, and its overexpression also inhibits the efficiency of reprogramming. Also, knockdown of Ythdf2/3 could upregulate TEAD2 targeted EMT-related gene expression, and promote cell migration, which indicates that knockdown of Ythdf2/3 could promote EMT process, which is known to repress reprogramming. Besides, by knocking down Tead2 or EMT related gene Tgfb1, the reduced reprogramming efficiency caused by Ythdf2/3 knockdown could be rescued. In addition, this study also revealed that the prolonged lifetime of somatic genes would inhibit the closure of somatic chromatin loci, which was not conducive to the opening of pluripotent gene related chromatin loci and reprogramming.
In this study, we used somatic reprogramming as a model to study the effects of the m6A binding protein YTHDF protein family on cell fate transition, which showed that YTHDF1/2/3 played a different role in reprogramming, and confirmed that YTHDF2 and YTHDF3 regulated somatic reprogramming synergistically through different RNA degradation pathways. This research result may provide insights into iPSC reprogramming at the posttranscriptional level.
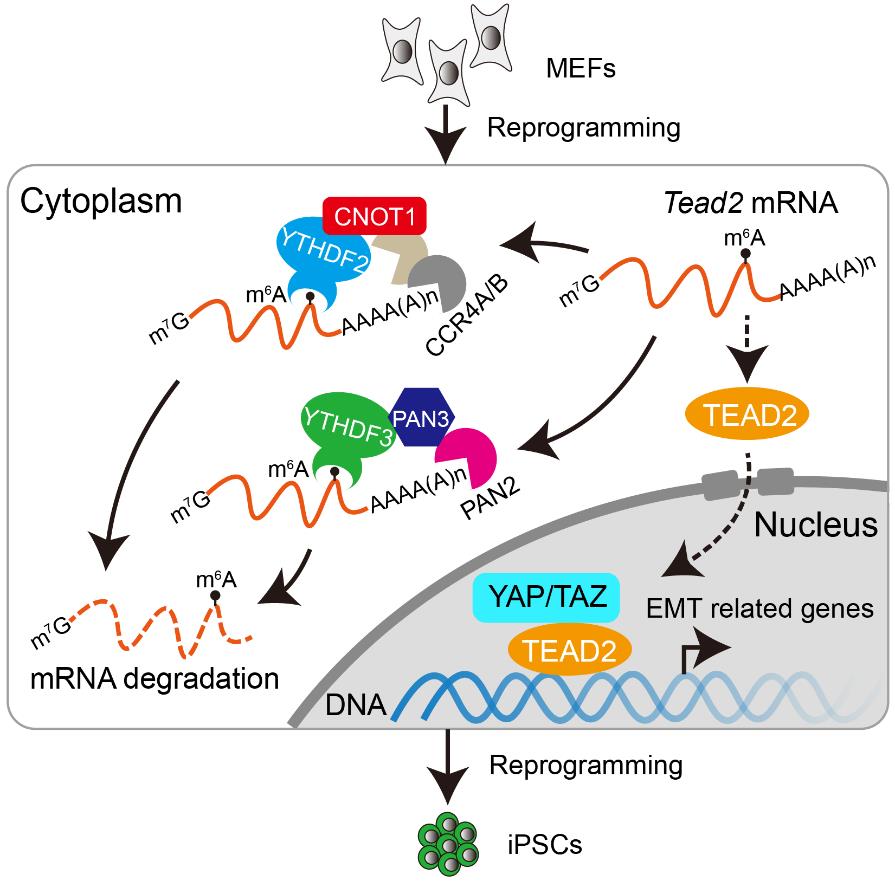
Figure 1. YTHDF2/3 regulated somatic reprogramming synergistically through different RNA degradation pathways.
Contact:
Chen Jiekai
Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences
E-mai:chen_jiekai@gibh.ac.cn