Researchers reported a new method for identifying ubiquitin interactions
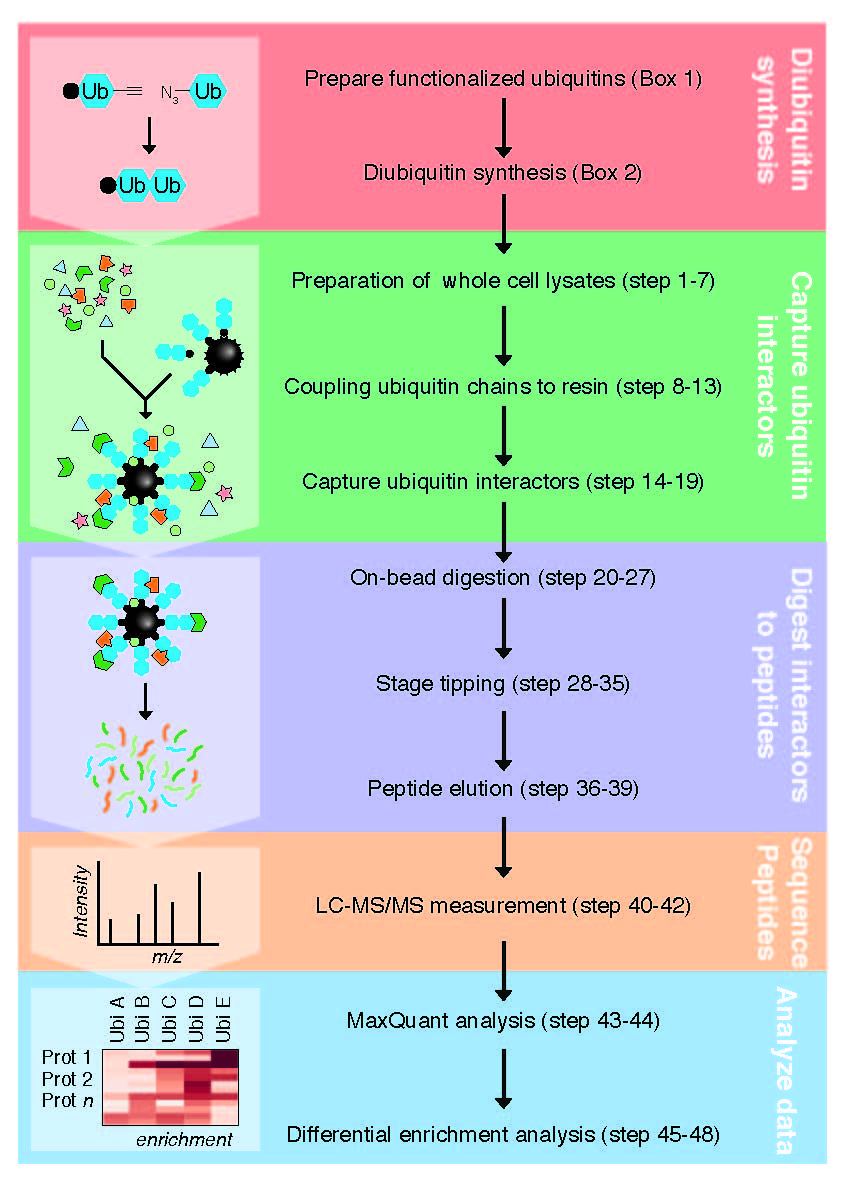
The covalent attachment of ubiquitin molecules to protein substrates is essential for almost all eukaryotic cellular processes. Current knowledge about ubiquitin-binding proteins and ubiquitin linkage-selective interactions is mostly based on case-by-case studies. To comprehensive identify ubiquitin interactors for all ubiquitin linkages in a relative physiological condition, Dr. Zhang Xiaofei from Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences and Dr Michiel Vermeulen from Radboud University, the Netherlands have recently reported a novel work flow called ubiquitin interactor affinity enrichment–mass spectrometry (UbIA-MS) to identify all ubiquitin linkages interactors. This paper has been published in Nature Protocols. UbIA-MS contains five stages: (i) chemical synthesis of ubiquitin precursors and click chemistry for the generation of biotinylated nonhydrolyzable diubiquitin baits, (ii) in vitro affinity purification of ubiquitin interactors, (iii) on-bead interactor digestion, (iv) liquid chromatography (LC)–MS/MS analysis and (v) data analysis to identify differentially enriched proteins. Typically, UbIA-MS allows the identification of dozens to hundreds of ubiquitin interactors from any type of cell lysate, and can be used to study cell type or stimulus-dependent ubiquitin interactions. Furthermore, this paper provided computational analysis tools are freely available as an open-source R software package, including a graphical interface. This R package is not restricted to UbIA-MS experiments but can be used to analyze different types of mass spectrometry experiments.