Polyomino: a high-precision single-cell and spatial omics integration algorithm
On October 22, 2025, Genome Research published online a paper from Professor CHEN Jiekai’s laboratory at the Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, entitled “Polyomino reconstructs spatial transcriptomic profiles with single-cell resolution via region-allocation method.” The paper was selected as the cover article for issue 11 of Genome Research. The study introduces a spatial integration algorithm, Polyomino, which efficiently and accurately reconstructs spatial transcriptomic maps at single-cell resolution.
Cells in living organisms do not exist in isolation. They communicate with each other according to their spatial positions to form intricate tissues and organs. Spatial transcriptomics is a key technology for revealing this cellular blueprint and has advanced rapidly in recent years. However, existing methods for integrating single-cell and spatial datasets often struggle to balance speed and accuracy when confronted with data at the scale of millions of cells, due to computational demands and noise.
The research team adapted the image-analysis concept of regions of interest (ROIs) and developed a multi-level region-allocation strategy to place cells spatially. Rather than assigning cells directly as in conventional approaches, Polyomino first partitions the spatial data into an array of tiles and constrains cell localization using multi-level regional cues (such as horizontal/vertical bands and spatial clustering). Fine-scale matching is then performed within each tile. This two-stage design dramatically reduces computational cost and improves robustness to common noise sources such as cell-segmentation errors and imbalance in cell-type proportions.
In a series of evaluations on both simulated and real data, Polyomino demonstrated outstanding performance:
a) High computational efficiency: Integrating roughly 15,000 single cells with nearly 20,000 spatial spots required only 141 seconds, 10 to 1,000 times faster than existing algorithms. Polyomino is currently the only method capable of integrating million-scale cells in a single run.
b) Accuracy and robustness: The method maintained high accuracy under various noise scenarios (e.g., segmentation errors, proportion biases). In validations using mouse embryos and cerebral cortex samples, Polyomino accurately reconstructed spatial distributions of cells.
c) Broad applicability: In analyses of colorectal cancer liver metastasis tissues, Polyomino revealed spatially distinct subpopulations of conventional dendritic cells (cDCs) between tumor and peritumoral regions and identified immune signaling networks associated with angiogenesis, offering new insights into the tumor microenvironment.
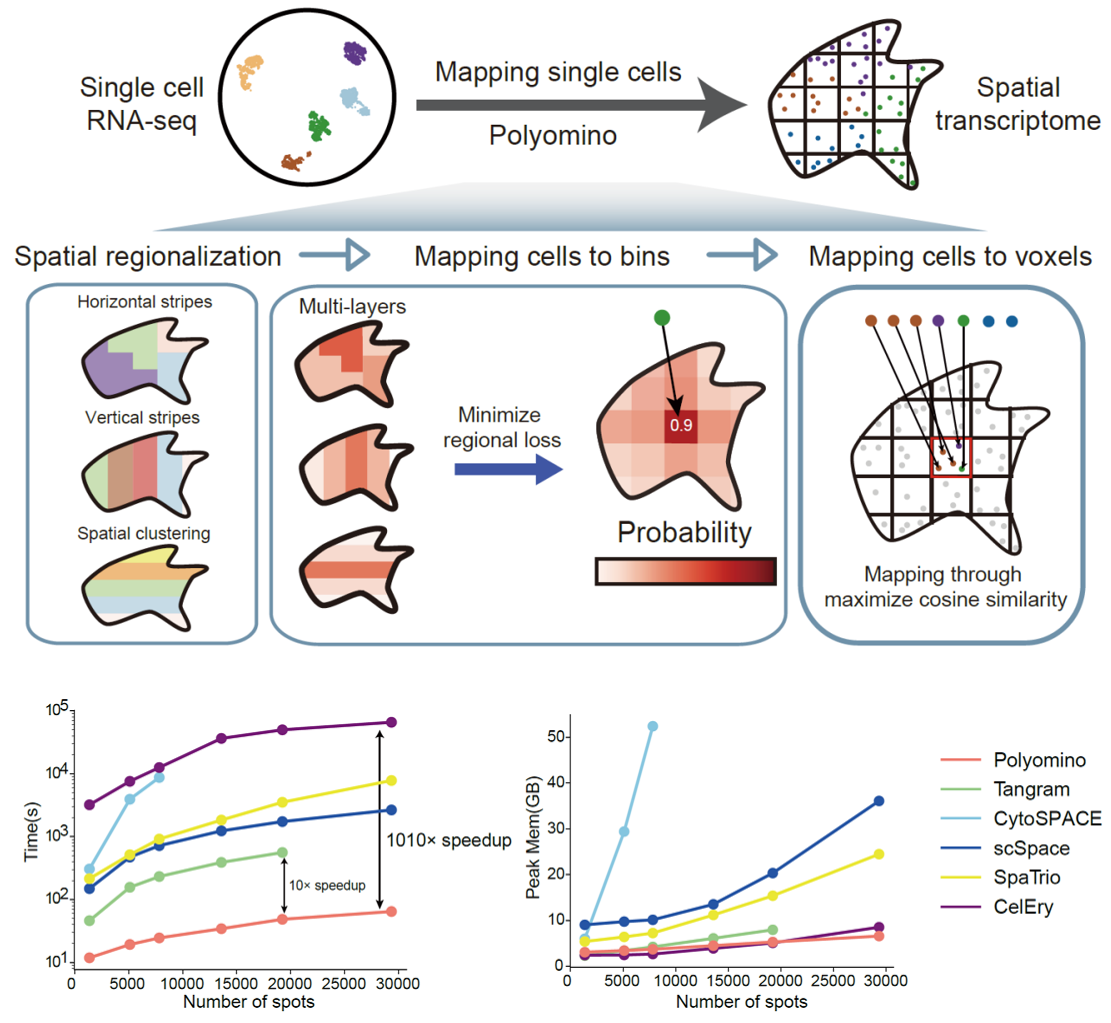
Polyomino’s multi-level region-allocation strategy enables single-cell spatial localization while substantially improving computational efficiency (Image by Prof. CHEN's team)
Overall, Polyomino overcomes computational bottlenecks in large-scale data integration and provides a higher-resolution tool for studying tissue development, disease mechanisms, and potential clinical applications. The authors anticipate that, as single-cell and spatial omics datasets continue to grow, Polyomino will become an important methodological foundation for building comprehensive cellular atlases and for dissecting organogenesis and pathological changes.
Dr. CAI Quanyou, Associate Investigator LIN Lihui from GIBH, CAS and Dr. LIU Xin from Guangdong Second Normal College are listed as co-first authors. Professor CHEN Jiekai and Associate Investigator LIN Lihui are co-corresponding authors.
This work was financially supported by the National Key R&D Program of China, the National Natural Science Foundation of China, a Major Project of Guangzhou National Laboratory, the Science and Technology Planning Project of Guangdong Province, China, and the Health@InnoHK program launched by the Innovation Technology Commission of the Hong Kong SAR, P.R. China.
Contact:
CHEN Jiekai, Ph.D., Principal Investigator;
Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, China, 510530.
Email: chen_jiekai@gibh.ac.cn
Attachment Download:
-
ContactCHEN Jiekai, Ph.D., Principal Investigatorchen_jiekai@gibh.ac.cn
-
Reference
-
Related Article