GIBH Researchers Launch New Single-Cell Multi-Omics Method for Comprehensive Analysis of Pancreatic Cancer Circulating Tumor Cells
Recently, Professor LIN Da at the Center for Cell Lineage Technology and Engineering, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, together with collaborators, published a research article in Nature Communications reporting the development of a new single-cell multi-omics technology — Uniform Chromosome Conformation Capture (Uni-C). This method enables simultaneous analysis within a single cell of large-scale genomic alterations (such as SVs, CNVs, and ecDNA), small-scale mutations (SNPs/INDELs), and 3D chromatin architecture, thus achieving high-resolution, multi-dimensional joint profiling.
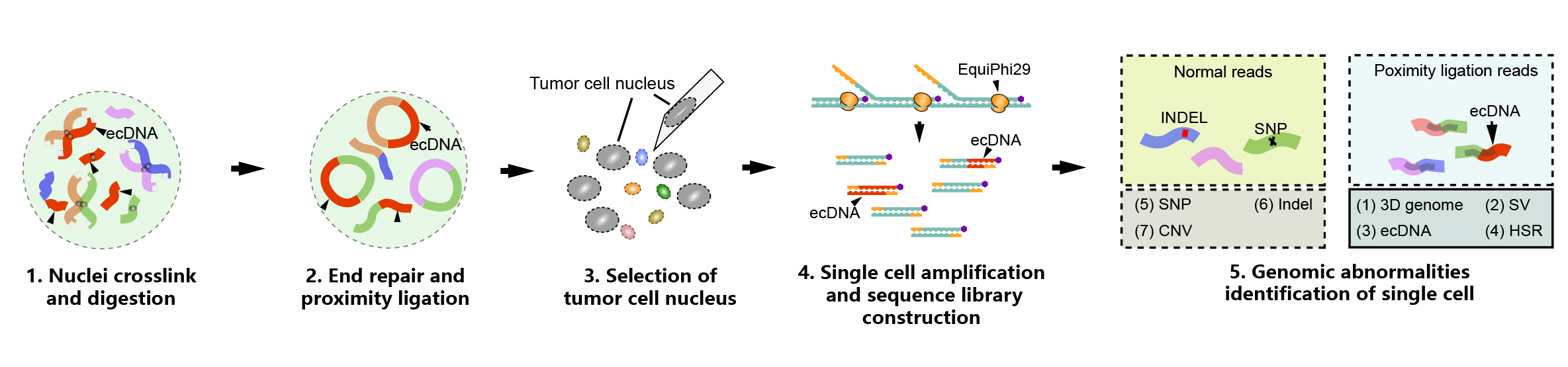
Figure: Workflow of Uni-C single-cell multi-omics library construction (Image by Prof. LIN's team)
Pancreatic cancer is characterized by pronounced heterogeneity and rapid progression, with clinical challenges in early detection and limited therapeutic targets. Circulating tumor cells (CTCs), which carry the complete genomic information of the primary tumor, have been considered a crucial window for non-invasive detection and dynamic monitoring. However, due to their scarcity and the difficulty of single-cell analysis, key enabling technologies are urgently needed.
Using Uni-C, the research team analyzed CTCs derived from pancreatic cancer. Remarkably, integration of data from only seven cells was sufficient to recover approximately 89% of SNPs/INDELs and 75% of structural variants, showing strong concordance with tumor tissue profiling. This validates the accuracy and representativeness of Uni-C in rare-cell contexts. Further analyses demonstrated that Uni-C can resolve the molecular structures of ecDNA in CTCs and reveal chromatin conformation changes across different cell-cycle stages — including A/B compartment reorganization and loss of higher-order structures — thereby providing new dimensions for evaluating CTC activity and cellular states.
Importantly, 3D chromatin organization not only carries spatial information but is also tightly linked to gene expression. Chromatin conformation data acquired by Uni-C at the single-cell level open up opportunities to explore potential connections between genomic alterations and transcriptional regulatory networks. For example, the folding states of chromatin across cell-cycle stages reflect transcriptional activity dynamics, highlighting the potential of Uni-C in dissecting mechanisms of tumor cell heterogeneity.
In terms of functional applications, Uni-C-derived mutation data were integrated into personalized neoantigen prediction. Synthetic peptide and in vivo experiments validated that several candidate neoantigens exhibited strong immunogenicity. When combined with immunotherapy, these neoantigens effectively suppressed tumor growth, demonstrating promising therapeutic potential.
This study represents the achievement of joint analysis of multiple types of genomic abnormalities and chromatin conformations in CTCs at the single-cell level, with the added ability to explore their functional significance in transcriptional regulation and immune responses. Uni-C provides powerful technical support for lineage tracing, target discovery, and personalized therapy of rare tumor cells, advancing the translational application of single-cell multi-omics in precision oncology.
Contacts:
LIN Da, Ph.D., Principal Investigator;
Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, China, 510530.
Email: lin_da@gibh.ac.cn
Attachment Download:
-
ContactLIN Da, Ph.D., Principal Investigatorlin_da@gibh.ac.cn
-
Reference
-
Related Article