Researchers Develop a Fluorosulfate Containing Enrichable and Latent Bioreactive Unnatural Amino Acid for Protein Interaction Research
Prof. TANG Shibing from Guangzhou Institutes of Biomedicine and Health (GIBH), the Chinese Academy of Sciences, in collaboration with Prof. YANG Bing of Zhejiang University and associate Prof. LIU Chao of Beihang University, published an article entitled with "Characterize direct protein interactions with enrichable, cleavable and latent bioreactive unnatural amino acids" on Nature Communications. They developed a fluorosulfate containing enrichable and latent bioreactive unnatural amino acid to characterize protein-protein interactions in live cells.
The identification of protein-protein interactions in live cell is important in the function study of proteins, and plays improtant roles in biomedical research. Proximity-enabled chemical cross-linkable unnatural amino acids (Uaas) have been used for the in situ identification of protein interactions in live cells via their ability to capture weak and transient protein interactions. Identification of cross-linked peptides with mass spectrometry is critical to ensure the specificity of interacting protein identification and reveal the protein interaction interfaces. However, due to the complexity of protein samples and data analysis process, it is challenging to perform high-throughput identification of Uaa-mediated cross-linked peptides in live cells. Enrichment of cross-linked peptides before mass spectrometry analysis can improve the identification efficiency.
In this study, researchers developed an fluorosulfate containing latent bioreactive unnatural amino acid eFSY, shorted for enrichable FluoroSulfate-L-tyrosine. eFSY can be incorporated into specific site of target proteins via genetic code expansion. The fluorosulfate of eFSY covalently cross-linked with tyrosine, histidine, or lysine in interaction proteins via a proximity-enabled sulfur-fluoride exchange (SuFEx) reaction. The alkyne group of eFSY can be biotinylated by copper-catalyzed azide-alkyne cycloaddition (CuAAC) and be used to enrich cross-linked peptides at the peptide level, thus improving the sensitivity of cross-linked peptides identification. In addition, researchers demonstrated that eFSY mediated cross-linking was fragmented in different patterns in mass spectrometry. While the sulfamate bonds formed by cross-linking with histidine and lysine are mass spectrometry cleavable, the cross-linking bond with tyrosine is not. To utilize this characteristic of eFSY cross-linking product, the cross-linking identification software AixUaa was developed. AixUaa is compatible with both cleavable and non-cleavable modes at the same time, which can precisely match the cross-linking peptides and sites, and increase the number of identified cross-linked peptides. Researchers systematically identified direct interactomes of Thioredoxin 1 (Trx1) and Selenoprotein M (SELM) with eFSY and AixUaa in Escherichia coli and mammalian cells separately.
In summary, the fluorosulfate containing enrichable and latent bioreactive unnatural amino acid eFSY and the cross-linking identification software AixUaa that can significantly improve the identification efficiency of cross-linking peptides and overcome some shortcomings of traditional chemical crosslinkable unnatural amino acids. It will be a promising method in protein-protein interaction study.
The research was supported by the National Natural Science Foundation of China, the National Key Research and Development Program of China, the Natural Science Foundation of Zhejiang Province, and the Basic Research Project of GIBH.
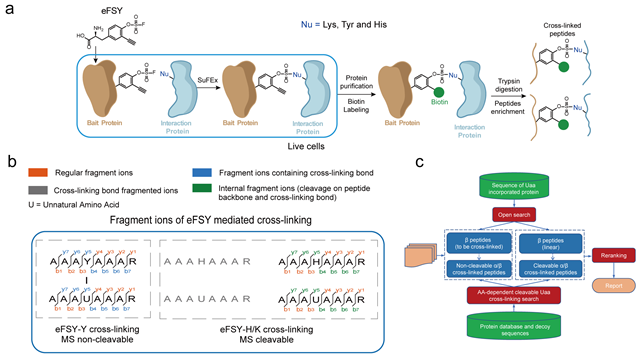
Caption: Flow chart for the identification of cross-linked peptides (a), fragmentation modes of cross-linked peptides (b) and development process of the cross-linking identification software AixUaa (c) for the study of protein interactions by eFSY, a novel fluorosulfate containing enrichable and latent bioreactive unnatural amino acid
Contacts:
Shibing Tang, Ph.D., Principal Investigator
Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences
Guangzhou, China, 510530
Email: tang_shibing@gibh.ac.cn
Attachment Download:
-
Contact
-
Reference
-
Related Article