Novel Spatial Multi-omics Technology Deciphers Spatiotemporal Lineage of Mouse Brain Development
Human and many other mammals are composed of billions of cells, which, although originating from the same fertilized egg, differ in types, developmental timing, and spatial distribution. But how are the identity and function of these cells determined still unknown? In general, they primarily depend on intrinsic and extrinsic factors. Intrinsic factors involve the interactions between the genome, epigenome, transcriptome, proteome, metabolome, and other components, centered around the central dogma. Extrinsic factors, on the other hand, can be regulated through interactions with neighboring cells.
Spatial omics technology not only effectively preserves the spatial location information of cells in situ but also captures the transcriptomic information within cells simultaneously. However, current spatial omics technology is mainly limited to single-omics. The spatial multi-omics technology that aims to simultaneously obtain multiple omics information within the same tissue is expected to facilitate the elucidation of the spatiotemporal lineage and the relevant regulatory mechanisms of human cells, thereby better serving the treatment of human aging and diseases.
On May 25, the research team led by Dr. PENG Guangdun from the Guangzhou Institutes of Biomedicine and Health (GIBH) of Chinese Academy of Sciences, published a research paper entitled"Simultaneous profiling of spatial gene expression and chromatin accessibility during mouse brain development" in Nature Methods. The paper reported a spatial multi-omics technology called Microfluidic indexing-based spatial assay for ATAC and RNA-sequencing (MISAR-seq). This technology utilizes a microfluidic chip-based targeted barcode delivery system to simultaneously capture the genomic information of both Assay for Transposase-Accessible Chromatin (ATAC) and RNA within cells while preserving their spatial location information. The researchers successfully applied this technology to investigate the mechanisms of embryonic mouse brain development, constructing a spatial multi-omics map for mouse brain development. Furthermore, they revealed the specific regulatory mechanisms of cis-regulatory elements involved in the formation of promoters and enhancers during brain development.
The researchers applied MISAR-seq experimental platform to study mouse brain development at various embryonic stages, including E11.0, E13.5, E15.5, and E18.5. The quality of the captured ATAC and RNA data from MISAR-seq was found to be comparable to the corresponding single-omics data, with each grid (5-20 cells) detecting approximately 2500-3500 genes. Integration analysis with single-cell data showed that the cell types identified by this technology matched well with the brain structure annotations of the corresponding single cells, indicating the robustness of MISAR-seq in capturing two types of omics data simultaneously.
By integrating and analyzing the mouse brain development data at different developmental stages containing both ATAC and RNA information, the researchers developed a bioinformatics tool that combined spatial constraints and image feature extraction. This tool improved the "horizontal" and "vertical" integration capabilities of the data between different omics at multiple time points, enabling the construction of a spatial multi-omics map for mouse brain development. The results demonstrated that both the ATAC and RNA data captured by MISAR-seq, either individually or together, could effectively delineate brain regions, and the clustering results were consistent with the annotations from the Allen Brain Atlas.
Using the obtained MISAR-seq data, the researchers also plotted the spatial and temporal trajectories of both omics during cortical development. They proposed a concept called the Domain of Enhancer Regulatory (DER), which reflected the strength of gene expression intensity and the number of connected enhancer peaks. Further analysis of enhancer and promoter regulatory networks revealed that the direct cascading regulation established by Pax6-Eomes-Tbr1 determins the sequential and region-specific gene expression during mouse cortical development.
In summary, the MISAR-seq technology not only achieved the simultaneous capture of ATAC and RNA information at the spatial level but also successfully constructed a spatial multi-omics map for mouse brain development, elucidating the specific regulatory mechanisms of promoters and enhancers in cortical development.
In the future, the researchers will conduct further optimization and expansion of MISAR-seq in spatial resolution and three-dimensional tissue analysis. They also anticipate its compatibility with additional omics information and the development of improved bioinformatics tools for spatial multi-omics gene regulatory network analysis, aiming to reveal the specific mechanisms determining cell identity and function in structurally complex tissues.
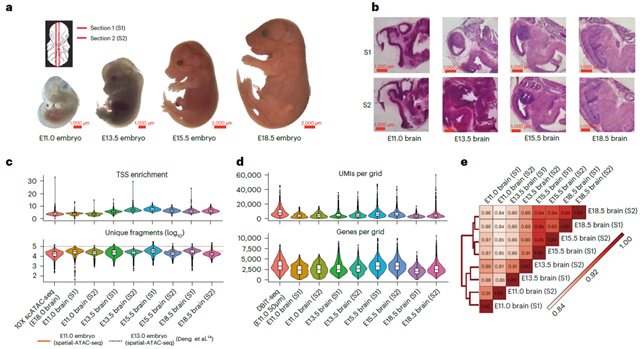
Fig. 1 Overview of MISAR-seq: data quality and validation. (Image by GIBH)
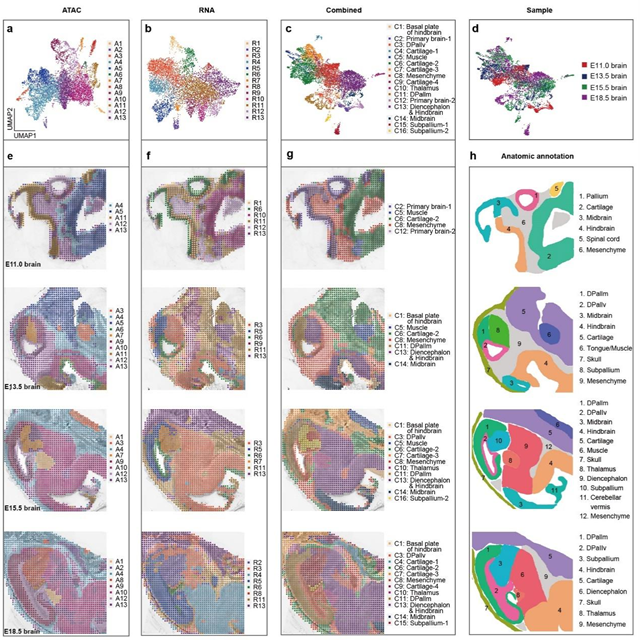
Fig. 2 Spatial chromatin accessibility, gene expression and combined mapping for mouse brain. (Image by GIBH)
Contacts:
Peng Guangdun,Ph.D, Principal Investigator
Guangzhou lnstitutes of Biomedicine and Health, Chinese Academy of SciencesGuangzhou, China, 510530
Email:peng_guangdun@gibh.ac.cn
Attachment Download:
-
Contact
-
Reference
-
Related Article