Researchers develop a novel gene base editor free of Cas9-dependent off-targeting
Recently, Prof. LAI Liangxue from Guangzhou Institutes of Biomedicine and Health (GIBH) of Chinese Academy of Sciences collaborating with Dr. ZOU Qingjian from Wuyi University, has developed a novel base editing system, TaC9-ABE, which is able to eliminate Cas9-dependent off-targets of adenine base editor (ABE).
This work was published in Cell Discovery on March 24, 2022.
Base editor enables single-nucleotide conversions without creating double-stranded DNA breaks. As an efficient and precise gene-editing tool, it can be used to generate gene-editing organisms and correct pathogenic point mutations for gene therapy.
ABE can specifically convert adenine (A) to guanine (G), two nucleotides of DNA, at the target sites. The canonical ABE contains a fusion protein composed of nCas9 and adenine deaminase and is guided by sgRNAs to create A-to-G conversion. However, sgRNAs tend to bind some sites with one or more mismatched nucleotides in genomes, thus resulting in off-target gene editing, which is the major safety concern in the generation of gene editing organisms and gene therapy for human diseases.
In the TaC9-ABE editing system of this study, nCas9 is guided to the target site by sgRNA, while the adenine deaminase is guided by transcription activator-like effector (TALE). Only when nCas9 is guided to the target site close to the TALE target to open the DNA duplex and nick the targeted strand, TALE-guided adenine deaminase can convert A to G on the on-target single strand. If nCas9 is guided to a wrong site (mismatched off-target site) by sgRNA, base editing is not generated due to the absence of deaminase. Similarly, when deaminase is guided to the wrong site by TALE, the base editing will not happen due to the absence of single-strand DNA substrates. Thus, such base editing system can completely rule out the possibility of Cas9-dependent off-target.
The results confirmed that TaC9-ABE maintained the high base editing efficiency and did not lead to any detectable genomic off-target modification at off-target sites. By contrast, half of the Cas9-dependent off-targets were observed in the canonical adenine base editor, ABE7.10.
“Our research further solves the safety issue of single base editing technology in biomedical applications, especially providing a safer and more efficient tool for the treatment of human genetic diseases caused by point mutations.” Prof. LAI says.
The study is sponsored by the Ministry of Science and Technology, the Chinese Academy of Sciences, the National Natural Science Foundation of China, Guangdong Provincial Government, and Guangzhou Municipal Government.
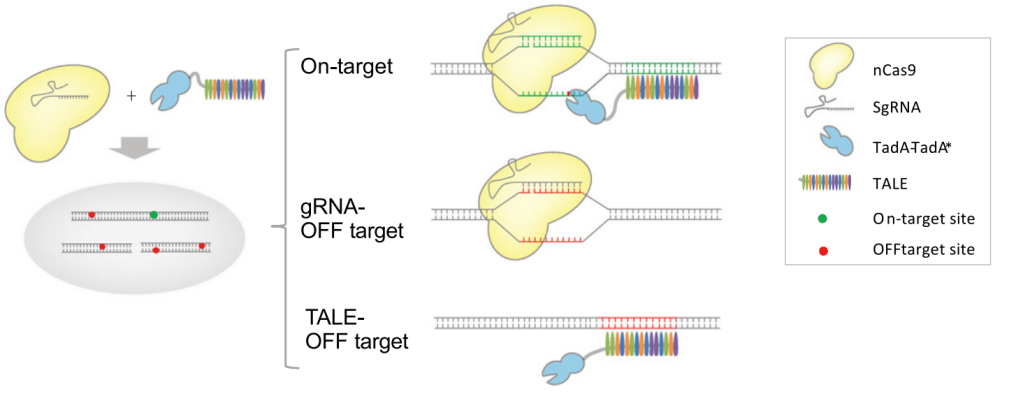
Schematic diagram of the base-editing principle of TaC9-ABE at the target and off-target sites. (Image by GIBH)
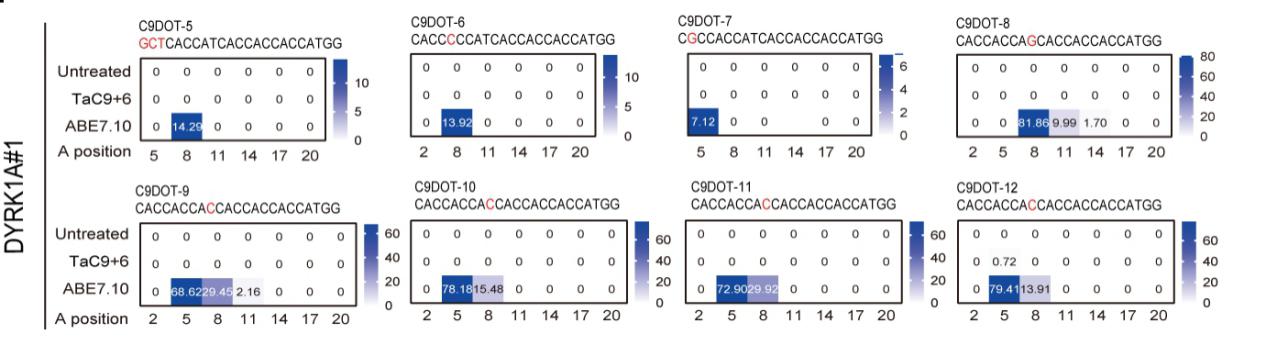
No Cas9-dependent off-targets were detected by TaC9-ABE. (Image by GIBH)
Contact
LAI Liangxue, Ph.D Principal Investigator
Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences
Guangzhou, China, 510530
Email: Lai_liangxue@gibh.ac.cn
Attachment Download:
-
Contact
-
Reference
-
Related Article