Scientists reveal the cooperative function of JMJD3 and KLF4 in somatic reprogramming
Ten years ago, PEI Duanqing, Miguel A. Esteban, WANG Tao, QIN Baoming and other team members--reported that vitamin C (Vc) greatly boosts the efficiency of both mouse and human somatic cell reprogramming. They demonstrated that Vc inhibits cell senescence and accelerates pluripotency gene activation. However, the detailed molecular mechanisms of the powerful effect of Vc was unclear. They noticed then that in biochemistry, Vc has been widely used as a reducing agent for assaying in vitro the activity of a panel of Fe2+ and ??ketoglutarate dependent dioxygenases, including the Jumonji family histone demethylases and TET DNA hydroxylases. Based on this property, they hypothesized that the effect of Vc in speeding up cell fate transitions in reprogramming, and probably also in other contexts, comes from these demethylases. Indeed, our later studies identified several such enzymes as Vc targets in reprogramming, including JHDM1B, KDM3/4 and TET1. Vc thus provided us with a unique and efficient tool to illuminate the epigenetic black box of cell fate transitions and, definitely, as they expected, other novel mechanisms awaited discovery.
As one of the most classical epigenetic markers in development and stem cell differentiation, H3K27me3 is deposited and removed genome-wide in a highly dynamic manner to ensure the right temporal and spatial changes in gene expression. Somatic cell reprogramming can be viewed largely as a reversion of development, and so is the change of H3K27me3 modification. For example, many pluripotency and epithelial genes are modified and silenced by H3K27me3 in somatic fibroblasts and their activation during reprogramming is accompanied by H3K27me3 demethylation. Based on this, theyhypothesized that in reprogramming, demethylation of H3K27me3 is a necessary and also rate-limiting step.
In mammalian, there are mainly two H3K27me3 demethylases: JMJD3 (KDM6B) and UTX (KDM6A). Even though highly similar in their catalytic domains, these two enzymes are distinct in other domains and functions. Two previous studies had revealed that whereas UTX is necessary for reprogramming, JMJD3 is detrimental. The inhibitory role of the latter was reported to come from induction of cell senescence and degradation of the pluripotency related factor PHF20. Noticeably, both studies were carried out in culture conditions without Vc. We thus asked whether these demethylases play different roles in reprogramming medium with Vc.
On October 8, Nature Communications journal published a research article titled "JMJD3 acts in tandem with KLF4 to facilitate reprogramming to pluripotency" from the laboratories of QIN Baoming and Miguel A. Esteban in the Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences.
In this study, they demonstrated that UTX is essential for reprogramming, as reported, while JMJD3 has two opposing effects. On one hand, consistent with the earlier study, we could observe that JMJD3 inhibits reprogramming in late-passage mouse embryonic fibroblasts (MEFs) through inducing the known pro-senescence target Ink4a/Arf, in a reprogramming independent manner. On the other hand, in early-passage (young) MEFs, JMJD3 boosts reprogramming efficiency and the effect is potentiated by Vc (see below the schematic). Mechanistically, in young MEFs, JMJD3 is recruited specifically by KLF4 to many epithelial and pluripotency gene loci and facilitates their activation. In addition to reprogramming to induced pluripotent stem cells, JMJD3 also enhances other KLF4-driven cell fate transitions.
In perspective, our discovery provides informative insight for a better understanding of the complex roles of JMJD3 and KLF4 in development, stem cell differentiation, as well as in physiological and disease-related contexts.
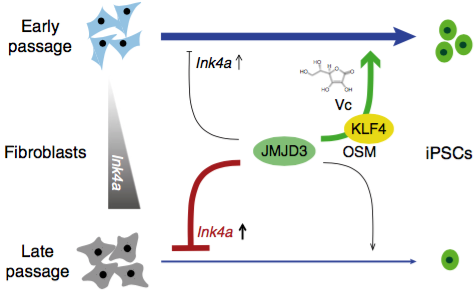
JMJD3 positively and negatively regulates reprogramming in early and late passage fibroblasts respectively (Image by QIN)
Contact:
Baoming Qin, Ph.D., Principal Investigator
Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences
E-mail: qin_baoming@gibh.ac.cn
Link:https://www.nature.com/articles/s41467-020-18900-z
Attachment Download:
-
Contact
-
Reference
-
Related Article