Hundreds of proteins have been identified as RNA binding proteins, which includes proteins associated with various diseases such as neural degeneration, immune system defects and cancer. This raised the necessity on systematically isolating RNA binding proteins. Current approaches for systematically characterizing the RNA binding proteins are mainly based on the capture of polyadenylated (polyA) RNAs, which fails to capture proteins interacting with non-polyA RNAs.
The research led by Miguel Esteban, Biliang Zhang and Xichen Bao from Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, has been published in Nature Methods entitled“Capturing the Interactome of Newly Transcribed RNA”. This methodology would not only provide a novel approach for systematically analyzing RNA-protein interaction in cells, but also shed light on the biological functions of genomic ‘dark matter’- long non-coding RNAs.
The researchers combined RNA labeling with characterization of bound proteome. Application of the methodology (RICK: capture of the newly transcribed RNA Interactome using ClicK chemistry) has successfully identified hundreds of novel RBPs with extensive binding to the previous neglected non-polyA RNAs. Besides, short labeling of RICK also captures proteins interacting with nascent RNAs, to understand the events coupled with nascent RNA production.
This work was funded by the National Key Research and Development Program of China, the National Natural Science Foundation of China, the Pearl River Science and Technology Nova Program of Guangzhou, the Youth Innovation Promotion Association of the Chinese Academy of Sciences and so on.
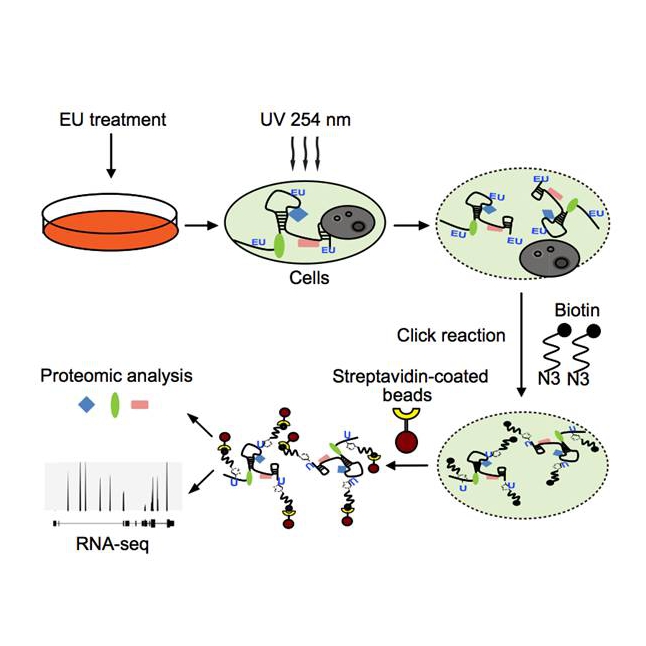
Flowchart of RICK methodology