GIBH scientists discovered that dynamically reorganized chromatin is the key for the reprogramming of somatic cells to pluripotent cells
During the progression from mouse embryonic fibroblasts (MEFs) to pluripotent stem cells (iPSCs), the cell undergoes a remodeling of chromatin architecture. Nucleosomes are the fundamental structural units of chromatin, consisting of approximately 146 base pairs of DNA wrapping around a histone octamer. Key epigenetic modificatons of chromatin structure include the positioning of nucleosomes and covalent modifications to histone tails, which regulate the expression of specific genes. Global maps of nucleosome positions in C. Elegans, S.cerevisiae and humans show that nucleosomes are not randomly distributed throughout the genome but are highly organized, particularly at TSSs. Critically, dynamic changes of nucleosome positions have also been observed during the differentiation of mouse ESCs to neural progenitors. However, understanding how nucleosome positioning and remodeling affects somatic cell reprogramming remains to be determined.
Research works carried out by Prof Hongjie Yao and his group have determined the genome-wide nucleosome coverage and histone methylation occupancy in MEFs, induced iPSCs and pre-iPSCs. Firstly, Prof Yao and his group have generated genome-wide maps of nucleosome occupancy and the distributions of active (H3K4me3) and repressive (H3K27me3 and H3K9me3) histone modifications. They investigated these marks during the conversion of MEFs to pre-iPSCs, and then to iPSCs via chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq) and carried out RNA-sequencing (RNA-Seq) to determine transcript abundance. Their study showed that iPSCs possess a more open chromatin state among the three cell lines. In addition, nucleosomes in pre-iPSCs are more phased than those in MEFs and iPSCs. Furthermore, cell type-specific genetic programs gradually shuts down, and pluripotency programs are activated. Nucleosome reorganization and histone methylation around TSSs are highly coordinated with gene activities during reprogramming. Bivalent promoters gradually increase and repressive promoters gradually decrease when reprogramming MEFs to pre-iPSCs, then to iPSCs. Active HCG promoters are characterized with nucleosome depletion at TSSs, while LCG promoters features an abundance of nucleosomes. Moreover, Prof Yao and his group showed that VC promotes nucleosome reorganization on specific genes during the fate transition from pre-iPSCs to iPSCs. Their results provide insights into the regulation of nucleosome repositioning and provide a basis for studying cell fate transitions during reprogramming.
Prof Yao and his team have provided a nucleosome remodeling roadmap from MEFs to iPSCs and conclude that the stage-specific organizations of nucleosomes and histone modifications represent an additional regulatory process that controls DNA access by regulatory factors to manipulate genes expression during somatic cell reprogramming. Their study strengthens the understanding of reprogramming process, which in turn benefits our understanding of cellular development and differentiation. The new picture of nucleosome reorganization should help us to understand new insights into the mechanisms involved in gene regulation and human diseases and therefore potentially leading to new therapeutic applications.
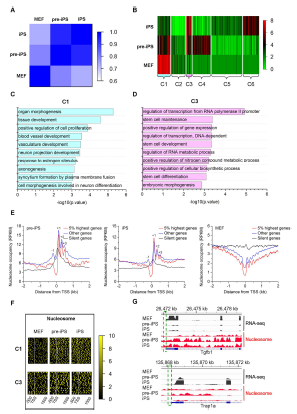
Correlations between gene expression and chromatin state during somatic cell reprogramming.
Attachment Download:
-
Contact
-
Reference
-
Related Article